Nitrogen Use Efficiency in Maize
Project overview:
Application of nitrogen fertilizer is required by all cereal plants to maximize yield. The increased input of N fertilizers and the related pollution have been concerns for both the producers and consumers. This concern is more pronounced in the developing countries. Therefore, identifying alleles that favor better Nitrogen Use Efficiency (NUE) and understanding the interaction among these alleles has been one of the goals of our lab. With this information, we might be able to increase corn yield under low N conditions.NUE is a complex trait and N uptake and N utilization are its major components (http://nitrogenes.cropsci.illinois.edu/). We employ different physiological, genetic, functional genomics approaches to dissect the molecular mechanisms that regulate NUE in maize. In addition, different experimental systems are currently being explored to associate changes in gene structure and expression with different genotypes, developmental stages, and environmental conditions.
The current project focuses on:
1-Discover genes associated with N response in maize
2-Cloning and functional analysis of key genes in N metabolic pathway
3-Understanding N/C balance during corn plant development and under
nitrogen stress.
Molecular dissection of N metabolism in the Illinois Protein Strains, B73 and Mo17
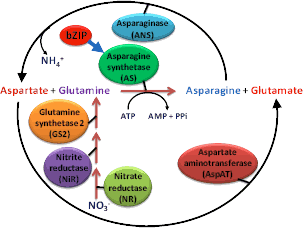 Figure
1: Asparagine metabolic pathway
Figure
1: Asparagine metabolic pathwayThe Illinois Protein Strains, B73, and Mo17 represent important genetic resources to investigate questions related to the physiological and molecular mechanisms that influence the ability of corn plants to assimilate, translocate, partition, and store carbon and nitrogen. We study differences in N related gene expression among these inbreds and correlate these to changes in grain composition and physiological responses to different rates of supplemental nitrogen under replicated field plot conditions.
Our objective is to determine regulatory networks of transcriptional activity that are associated with amino acid metabolism with particular emphasis on asparaginemetabolism (Fig 1) as a main system of N storage and transport in plants. This research promises to reveal novel approaches for improving NUE.
Association of allelic and expression variations in N metabolism
related genes with NUE phenotypes in diverse maize germplasm.
We characterize allelic variation in key structural as well as regulatory
genes from a broader array of maize germplasmsuch as, the founders of
the Nested Association Mapping (NAM, Yu et al., 2008), the inbred lines
that have recently become publicly available through expired Plant Variety
Protection Act, a number of tropical varieties and some select elite inbreds.
Then we investigate the association of allelic combinations with the accumulation
of key amino acids (eg. Asnand Gln) as metabolic indicators of N utilization.
Genetic mapping of quantitative trait loci (QTL) controlling
maize NUE and its component traits.
We use B73 x Mo17 hybrid, hybrids derived from crosses of the intermatedB73
x Mo17 recombinant inbred (IBMRI) lines with the Illinois High Protein1
inbred (IHP1), which represents the known genetic extreme for N uptake.
The IBMRI x IHP1 population is being phenotypedextensively for NUE traits,
amino acid profiles, and mRNA expression differences under different ialN
availability, which will identify quantitative trait loci (QTL) and expression
QTL (eQTL) controlling N response traits. Fine-mapping is in progress
to discover the genes underlying the QTL.
People
Jayanand BodduFarag Ibraheem
Yuhe Liu
